Cloning and Expression of Spider Dragline Silk Protein Gene in Escherichia coli and Eukaryotic Cells 
2. School of Agriculture, Eastern Liaoning University, Dandong, 118001, P.R. China
 Author
Author  Correspondence author
Correspondence author
Molecular Entomology, 2011, Vol. 2, No. 1 doi: 10.5376/me.2011.02.0001
Received: 19 Nov., 2011 Accepted: 26 Dec., 2011 Published: 28 Dec., 2011
Liu et al., 2011, Cloning and Expression of Spider Dragline Silk Protein Gene in Escherichia coli and Eukaryotic Cell, Genomics and Applied Biology, 30(1): 16-20 (doi: 10.3969/gab.030.000016)
With the extreme tensile strength and toughness, spider silk has a high application value in a wide range of medical and industrial. However, the application of spider silk was limited because of its non-domestication. So, we tried to obtain the spider silk protein by genetic engineering. In this study, the dragline silk gene (ASP), a 837 bp fragment was cloned from the genome of spider (Araneus ventricosus) by Nested PCR. And then it was ligated into pGEX-6p-1 prokaryotic expression vector and pGFP-N2 eukaryotic expression vector, named as pASG and pASN, respectively. The fusion protein GST-ASP was successfully expressed in pASG and characterized by Western blotting with anti-GST antibodies after induced at 16℃ for 24 h in E. coli. The fusion proteins ASP-GFP was also successfully expressed in pASN by the detection of green fluorescence after transfected in insect sf9 cells for 8 h, which indicated that the ASP gene was correctly expressed in E. coli and eukaryotic cells, respectively. This study provides a beneficial attempt to develop the pathway for yielding spider silk protein by genetic engineering.
Orb-web weaving spiders have 7 different highly specialized glands which can synthesize and secrete the spider silk, and each gland secretes the silk with different mechanical properties and functions respectively (Xu and Lewis, 1990). Dragline silk, used as the lifeline and spider silk frame, is one of the strongest silks in present studies. The extreme tensile strength of dragline silk is triple at the aramid fibers and five times than steel, therefore, it usually can be used in military affairs, industry, and medicine. Unfortunately, the dragline silk can not be obtained in large quantities from spiders, because of the inability of non-domestication of spider. Researchers have attempted to produce silk protein by genetic engineering (Hu et al., 2003). And it was proved that genetic engineering may be the only way to obtain natural spider silk protein.
As a result of abundance repetition sequence and high GC content in silk gene, clone and expression of spider dragline silk was very difficult. The recombinant dragline silk produced in E. coli was not apparent from crude protein extracted by SDS-PAGE, only detected in crude lysate by Western blotting (Fahnestock and Bedzyk, 1997; Arcidiacono et al., 1998). Kane (1995) thought that clusters of rare codons created most translational errors, although simply the presence of a little of these codons could introduce translational errors as well. Given this situation, one would predict that translational problems with an abundant mRNA species containing an excess of rare low tRNA codons. Ma et al (2007) found that recombinant proteins were synthesized at a very low speed in a optimized spider dragline silk proteins expression research. Similar spider dragline silk proteins expressed in mammalian cell were weaved into fibers. Howerer, the properties of them are far less than natural spider silks’ (Lazaris et al., 2002).
In this paper, we cloned the partial gene fragment of the spider protein from Araneus ventricosus genome by nested PCR, and successfully expressed it in E. coli and eukaryotic cells respectively. The results might lay a foundation for the study of production of artificial spider silk fiber.
1 Results and Analysis
1.1 cloning of partial gene of spider dragline silk protein and construction of expression vector
Major ampullae gland was dissected from spider (Araneus ventricosus) abdomens, and genome DNA was isolated. Results of agarose electrophoresis showed that the genome DNA sample present high integrality and purity (Figure 1).
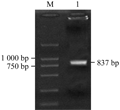
We obtained two DNA fragments of dragline silk by utilizing two nested PCR primers from the genomic DNA of Major ampullae silk gland, named as PCRA about 1 617 bp (GenBank accession No. DQ536503) and PCRB about 1 191 bp (GenBank accession No. DQ536504), respectively. After sequencing, the DNA sequences of PCRA and PCRB were aligned to the published partial cDNAs, the results showed that both fragments comprised exon and intron regions of dragline silk gene. The exon 1 (in PCRA), exon 2 and exon 3 (in PCRB) were spliced and then formed an 834 bp DNA fragment, named as ASP (GenBank accession No. DQ512478) (Figure 2; Figure 3).
The ASP fragment was digested with BamHâ… and EcoRâ… , EcoRâ… and Bglâ…¡, respectively. The products were ligated into vectors pGEX-6p-1 and pGFP-N2, which were dealt with the same enzymes. A fragment about 837 bp was identified in both recombinant plasmids by double enzyme digestion, we named the prokaryotic and eukaryotic expression as pASG and pASN, respectively.
1.2 Induced expression of pASG in E. coli
The prokaryotic expression vector pASG which contained ASP and GST was induced with 1 mmol/L isopropyl b-D-thiogalactoside (IPTG) for 24 h at 16℃ and 37℃ respectively. The cells were harvested by centrifugation and identified on SDS-PAGE. Only induced at 16℃, the fusion protein (58 kD) was apparent on the SDS-PAGE gel. The fusion protein was also detected by Western blotting using anti-GST antibodies. The results displayed that the specific bands in expression production were observed, which indicated that the fusion protein contained partial spider silk gene fragment has been expressed in E. coli (Figure 4).
1.3 Detection of transient expression of pASN in sf9 cells
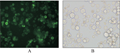
Sf9 cells were transfected with eukaryotic expression vector pASN containing ASP and GFP for 48 h to express the fusion proteins. GFP was observed by fluorescence microscopy (Figure 5), which showed that the ASP and GFP fusion gene can be expressed in the sf9 cells.
2 Discussion
Dragline silk renowned as a kind of crude high performance fibers, is attractive to biochemists and biomaterial scientists to study the relationship between protein design and function.
But it was not successful to manufacture the recombinant proteins by genetic engineering and the repetitive nature of these genes have contributed, at least in part, to the difficulties in their manipulation, which also existed in our studies.
The 834 bp ASP insert was a partial sequence of the published AVF, and both of them are from the 3’ end of Araneus ventricosus gene. The PCRA and PCRB, which were interspersed with introns, had areas of almost perfect homology to the published sequences of other spiders.
The 5' end of the PCRA was a intron about 1 420 bp in length and the 3' end was a repetitive exon (exon 1) which encoded two amino acid motifs: GPGG (X) n and GGX (G:Gly; P: Pro; X: other). The PCRB contained two exons, one (exon 2) was repetitive and the other (exon 3) contained the repetitive region and non-repetitive region. Craig et al (2000) reported the intron sequences were more homology within a species than that of the the exons, which indicated that the genome was similar in the species of the spiders.
In our studies, three exons were spliced and formed an 834 bp DNA fragment, a partial of the 3' end of Araneus ventricosus gene, which was proved can express in prokaryotic and eukaryotic systems. But the fusion protein was expressed in a high quantity only induced at 16℃.
Previous studies proved that ligated the dragline silk gene into prokaryotes had had some difficulties that the recombinant spider silk proteins were not visible from crude protein extracts by SDS-PAGE due to high GC content. The fusion protein was detected by SDS-PAGE in the crude lysate induced at 16℃, but not at 37℃. The reason is not apparent, we can only speculate that there are many low usage codons of the dragline silk genes in E. coli, but perhaps not for its high GC content. In our research, there are 16 codons of CCA, 5 codons of CCC, and 6 codons of AGU in the ASP sequence, which are rarely used in E. coli. Thus, when the recombinant spider silk proteins of ASP were induced at 37℃, the production of the fusion protein was so immediate and quick that the aminoacyl-tRNA can’t effectively identify the infrequent codons to translate the DNA into protein. When the fusion protein was induced at 16℃, the proteins of the E. coli were produced at very low speed, some aminoacyl-tRNAs could identify the rarely codons to express full-length protein.
3 Materials and Methods
3.1 Materials
Araneus ventricosus was collected from the campus of Dalian University of Technology in summer of 2008.
3.2 Instruments and Reagents
CK40 inverted microscope was purchased from Japan Olympus Corporation; Heraceâ…¡ CO2 incubator was purchased from America Kendro; Extreme ultraviolet spectroscopy was purchased Beijing Rayleigh Analytical Instrument Corporation.
Protease K was bought from Tiangen Biotech (Beijing) Co. LTD, and pmd18-T Vector, Ex Taq, and T4-DNA ligase were bought from Takara Biotechnology (Dalian) Co. LTD. Grace's culture medium of insect cells was bought from GIBCO, Cellfectin Reagent was purchased from Invitrogen. QIAprep Spin Miniprep Kit was purchased from Qiagen. DH5α and BL21 (DE3), Vector pGEX-6p-1 and pGFP-N2 were preserved in our experiment.
3.3 Gene clone and expression vector construction
Major ampullate silk glands were dissected and frozen immediately in liquid nitrogen. The glands were then crushed in a mortar and pestle under liquid nitrogen. DNA was isolated with DNA extraction buffer (10 mmol/L Tris-Hcl pH 8.0, 100 mmol/L EDTA, 0.5% SDS) at 50℃ for 4 h, and then Protease K was added to a terminal concentration 2 mg/mL at 37℃ for 2 h. After extracted with the same volume of phenol for twice and phenol/chloroform (1:1) for twice, the supernatant was precipitated with isopropanol and the precipitate was dried and dissolved in 400 μL TE. Crude extraction were extracted again with phenol, phenol/chloroform and chloroform respectively, precipitated with absolute alcohol, dried and dissolved in 200 μL TE. RNase was added to make the terminal concentration of 50 μg/mL, and was incubated at 37℃ for 30 min. Validated by agarose gel electrophoresis and extreme ultraviolet spectroscopy, DNA was conserved at -20℃.
According to AvMaSp1 gene sequence (GenBank accession No.: AY177203) of Araneus ventricosus dragline silk protein (Ren et al., 2002), two pairs of primers were designed to amplify the dragline silk gene by nested PCR (forward primerâ… , 5'-GGTGGACCAGGAGCTGGTG-3'; forward primerâ…¡, 5'-CCCGGGCCAGGAGGTATTTACGGACC-3'; Reverse primerâ… , 5'-GCTTGCATAATGGTAATTTATTA-3', Reverse primerâ…¡, 5'-GGTACCATATCCAAAGCATCTTCGGAC-3') from genomic DNA. With these two pairs of primers, PCR was performed by the template, totle DNA of major ampullae silk glands. The reaction system is 10 μmol/L, forward and reverse primers each 1 μL, DNA 1 μL, 2.5 mmol/L dNTP 4 μL, 10×Buffer 5.0 μL, Ex Taq 0.5 μL and ddH2O 37.5 μL. PCR procedure is 94℃ 50 s, 50℃ 1 min, 72℃ 2 min, 35 cycles, 72℃ extending 10 min. After 1% agarose gel electrophoresis, PCRA and PCRB were validated by EB.
To delete the introns, 3 pairs of primers were designed: F1 (gatcccgggggaggaccaggtgcaccagga), R1 (gtcctccagcgccactgacaactacagaagaa); F2 (agtggcgctggaggacccagtggaccaggagctggtg), R2 (accagctccacctcctggtccgaaagctccacct); F3 (ttcggaccaggaggtggagctggtggtccaggag), R3 (gatggtaccatatccaaagcatcttcggac). And PCRG (ASP) was formed by primers’ joint (Figure 6). PCR products were separated with the gel electrophoresis, the purification DNA, recovered by DNA Gel Extraction Kit, was ligated into pmd18-T vector. The recombinant vector was transformed into E. coli DH5α cells, and positive clone were chosen to extract plasmid and sequenced. The right fragment of ASP was inserted into pGEX-6p-1 (pASG) at BamHâ… and EcoRâ… sites, and pGFP-N2 (pASN) at EcoRâ… and Bglâ…¡sites. The recombinant plasmids were identified to obtain a 834 bp fragment by double enzyme digestion.
3.4 Expression of pASG in E. coli
The recombinant plasmid pASG was transformed into BL21 (DE3) E. coli competent cells. Positive clones were selected to culture by induced with 0.4 mmol/L isopropyl b-D-thiogalactoside (IPTG), centrifuged to haravest the cells. After 30 min (working power 400 W, operation time 10 s, distance 10 s) ultrasonic crash and centrifugation. The supernatants were identified on SDS-PAGE. The fusion proteins were detected with anti-GST antibodies by Western blotting through transferring the proteins onto a polyvinylidene difluoride (PVDF) membrane.
3.5 Expression of pASN in insect sf9 cells
Insect sf9 cells were cultured in Grace’s cell culture medium (GIBCO) with 10% FBS (TBD) at 27℃. The exponentially growing sf9 cells were inoculated in 24-well cell culture clusters, and transformed the recombinant plasmids using Cellfectin Reagent according to the procedures of specification. The cells were observed by fluorescence microscopy after 48 h post-transfection.
Authors' contributions
DML and WLL conceived the experimental design and conducted the data analyses, and wrote the manuscript. FW took an active part in experiment. All authors have read and approved the manuscript.
Acknowledgments
We thank two anonymous reviewers for their strict criticism on this paper.
Reference
Arcidiacono S., Mello C., Kaplan D., Cheley S., and Bayley H., 1998, Purification and characterization of recombinant spider silk expressed in Escherichia coli, Appl. Microbiol. Biotechnol., 49(1): 31-38 http://dx.doi.org/10.1007/s002530051133 PMid:9487707
Craig C.L., Riekel C., Herberstein M.E., Weber R.S., Kaplan D., and Pierce N.E., 2000, Evidence for diet effects on the composition of silk proteins produced by spiders, Mol. Biol. Evol, 17(12): 1904-1913 PMid:11110907
Fahnestock S.R., and Bedzyk L.A., 1997, Production of synthetic spider dragline silk protein in Pichia pastoris, Appl. Microbiol. Biotechnol., 47(1): 33-39 http://dx.doi.org/10.1007/s002530050884 PMid:9035408
Hu Q.L., Li X.D., and Shen J.C., 2003, Progress in structure biomimetic materials, Cailiao Yanjiu Xuebao (Chinese Journal of Materials Research), 17(4): 337-344
Kane J.F., 1995, Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coil, Current Opinion in Biotechnology, 6(5): 494-500 http://dx.doi.org/10.1016/0958-1669(95)80082-4
Lazaris A., Arcidiacono S., Huang Y., Zhou J.F., Duguay F., Chretien N., Welsh E.A., Soares J.W., and Karatzas C.N., 2002, Spider silk fibers spun from soluble recombinant silk produced in mammalian cells, Science, 295(5554): 472-476 http://dx.doi.org/10.1126/science.1065780 PMid:11799236
Ma H.W., Zhang L.S., Zheng W., Zhang Y.L., and Zhang Y.J., 2007, Exogenous alanine enhances the expression of spidro in cDNA of spider dragline silk protein in Escherichia coli, Zhongguo Shengwugongcheng Zazhi (China Biotechnology), 27(1): 47-51
Ren H.L., Liu Z.S., Pan F.G., Zhao J., and Li Y.S, 2002, Construction of Araneus ventuicosus major ampullate gland cDNA library and screening spider silk gene, Shengwu Jishu (Biotechnology), 12(5): 1-3
Xu M., and Lewis R.V., 1990, Structure of a protein super fiber: Spider dragline silk, Proc. Natl. Acad. Sci., USA, 87(18): 7120-7124 http://dx.doi.org/10.1073/pnas.87.18.7120
. PDF(307KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Danmei Liu
. Fen Wang
. Wenli Li
Related articles
. Dragline silk gene
. Gene cloning
. Prokaryotic expression
. Eukaryotic expression
Tools
. Email to a friend
. Post a comment