The Evaluation and Utilization of New Genes for Brown Planthopper Resistance in Common Wild Rice (Oryza rufipogon Griff.) 
2. Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007
3. Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007
4. China National Hybrid Rice Research Center, Changsha, 410125
5. Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007
6. Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
1 Guangxi Crop Genetic Improvement and Biotechnology Lab, Guangxi Academy of Agricultural Sciences, Nanning, 530007 2 Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 3 Agro-information Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007 4 China National Hybrid Rice Research Center, Changsha, 410125 5 Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, 530007; 6 Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization, Gusngxi University, Nanning, 530005
 Author
Author  Correspondence author
Correspondence author
Molecular Entomology, 2010, Vol. 1, No. 1 doi: 10.5376/me.2010.01.0001
Received: 07 Sep., 2010 Accepted: 12 Oct., 2010 Published: 26 Nov., 2010
Li et al 2006, The Evaluation and Utilization of New Genes for Brown Planthopper Resistance in Common Wild Rice (Oryza rufipogon Griff.), Molecular Plant Breeding, 4(3): 365-371
Background
Brown planthopper (BPH, Nilaparvata lugens St!l) is one of the most serious pests of rice in China and the world. More than 25% of the total chemicals applied in rice field are for control of this pest in China, which are expensive, environment unfriendly and injurious to human health due to their residual toxicity (Li et al.,1997). Breeding BPH-resistant varieties is therefore, the most economic and effective way to control this insect. BPH resistance is known to be in monogenic ordigenic inheritance in most resistance sources. More than 17 BPH resistance genes and some QTL loci have been identified in the cultivated and wild species of rice. Among them, more than 10 resistance genes have been mapped by molecular markers. Higher degree and broader spectrum of resistance present in cultivar Ptb33is conferred by bph2 and Bph3 together. (Sidhu and Khush, 1978; Khush et al., 1985;Kabir and Khush, 1988; Nomoto et al., 1989;Hirabayashi and Ogawa,1995; Kinoshita, 1995; Li et al., 1997ï¼›Hirabayashi etal., 1998; Jeon et al., 1999; Kawaguchi et al., 2001; Liu et al., 2001; Murata et al., 1997; 1998a; 1998b; 2001;Renganayaki et al., 2002; Yang et al., 2002; 2004;2005; Wu et al., 2005). Among the resistance genes,Bph1, bph2 and Bph3 have been used extensively in the breeding program. However, the varieties with Bph1and bph2 genes have lost their resistance in many rice growing regions due to change of BPH biotypes (Manisegaranet al., 1993; Medina et al., 1996). Therefore, it is important to identify new sources of high and broad spectrum resistance for rice breeding against BPH biotypese specially the virulent biotypes. The present report is undertaken to evaluate new resistance genes from the Guangxi wild rice species (Oryza rufipogon Griff.), and to use the genes against BPH biotypes.
1 The Evaluation for the Resistant Sources from Wild Rice
One thousand two hundred and fourteen entries of common wild rice (Oryza rufipogon Griff.) were collected from various regions in Guangxi where the wild rice was widely distributing and screened for their BPH resistance in the past 15 years. Only 30 (occupy 2.5%) of them were presented the resistance with resistant scores from 1 to 5, i.e. HR-highly resistant (scale1.0~2.0), R-resistant (scale 2.1~4.0), MR-moderately resistant (scale 4.1~6.0), MS-moderately susceptible (scale 6.1~8.0), S-highly susceptible (scale 8.1~9.0), while the other were susceptible with scores from 7 to 9. This result indicated that the frequency of BPH resistant resources in the wild species is very low, and it was possible to get new resistant sources/genes from the wild rice materials (Table 1).
|
Table 1 Resistance of Oryza rufipogon resources to Nanning local population of brown planthopper (BPH)
|
The resistant sources were collected from the 23 of46 counties of wild species habitat in the Guangxi. There was no evidence that the resistance sources accumulated in one site. Whereas, it seemed that the resistance resources was commonly present in the wild populations in certain frequency.
2 The Broad Spectrum of Resistance to the BPH Biotypes
Six BPH biotypes were used to find out their pathogenic reaction against the wild rice. Among them, biotype 1 and 2 were collected from Nanning in Southern China, and the Bangladesh biotype was the predominant type in Bengal regions of South Asia, and the Mekong biotype, Cuulong biotype and Pantnagar bio-366type were collected from Mekong and Cuulong deltas in southern Vietnam Pantnagar in the northern India respectively. The biotype 2 was most widely spreading inrice growing regions, while the Cuulong and Pantnagar biotypes were highly virulent (Pathak and Lal, 1976; Liet al., 1997). The results showed that the 6 wild rice germplasms used in the study were resistant to all of the5 or 6 BPH biotypes (Table 2). As compared with the tester varieties each having specific reaction to BPH biotypes, the wild rice germplasms were resistant to all the biotypes used in this study. Such kind of broad spectrum BPH resistance would be very valuable for breeding resistant varieties.
3 The Inher itance of BPH Resistance
The inheritance of BPH resistance in the wild materials was studied by crossing susceptible parent TN1 (P1) with the resistant parent 94-42-5-1 (P2) and other corresponding combinations and progenies. Parents, F1, F2, BC1P1 and BC1P2 populations were infested with Cuulong and Pantnagar biotypes.
All the F1 plants of the cross were susceptible and the plants in F2 population segregated in the ratio of 1 resistant: 3 susceptible suggesting dominance of susceptibility over resistance and the resistance being controlled by one pair of recessive gene. This result was further confirmed by the segregation in F3 lines into 1 resistant: 2 segregating: 1 susceptible. All the F3 lines derived from F2 resistant plants were uniformly resistant. TheBC1 progenies from backcross between F1 and susceptibleTN1 parent were all susceptible, whereas the progenies from testcross between F1 and resistant 94-42-5-1parent gave segregation ratio of 1 resistant: 1 susceptible plants (Table 3). Whereas, all the F1 plants of the cross were susceptible and F2 population segregated into 1 resistant: 15 susceptible plants. The test cross of F1with resistant parent 94-42-5-1 resulted in genetic segregation in the ratio of 1 resistant: 3 susceptible plants (Table 4).The inheritance of the resistance in 94-42-5-1 to the BPH biotype 2 also presented recessive in F1 plants and digenic segregation (1R:15S) in F2population. Screening of the other F2 populations (1R:15S) derived from different resistant sources (BPH94- 42-5-1, BPH2173,BPH2175, BPH 2182, BPH2183, BPH 2184, BPH2192,BPH 2195, BPH2200, BPH 2205) showed all the resistance was recessive digenic inheritance to the biotypes 2and Cuulong indicating two pair of resistance genes present in these materials.
|
Table 2 Resistance reaction of nucleus germplasm in O. rufipogon and O.sativa
|
|
Table 3 The genetic analysis of resistance in 94-42-5-1 against BPH Pantnagar biotype
|
|
Table 4 The genetic analysis of resistance of 94-42-5-1 against BPH Cuulong biotype
|
These results showed two pairs of independent recessive genes with duplicate interaction controlling resistance in 94-42-5-1. It suggested different genetic mechanism in the host against different BPH biotypes.
4 Molecular Mapping of the BPH Resistance Genes
The TC1F2 population from the cross between susceptible parent TN1 and resistant donor 2 183 was used as the mapping population. Plant resistance was evaluated by infestation with biotype 2. A total of 257 extreme resistant and susceptible plants were used for the molecular mapping of the resistance genes with 239 pairs of rice genome-wise SSR primers selected on the basis of their polymorphisms between parents. The genetic distances (centimorgan, cM) between resistance loci and markers were determined using Mapmaker/expV3.0 based on the segregation data in the BC2F2 mapping population ( LOD≥3.0).
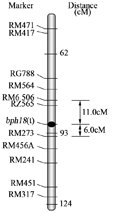
A total of 257 extreme resistant and susceptible plants were mapped with the marker for the resistance genes. The mapping showed three SSR markers, RM273, RM6506 and RM252, located in the middle of long arm of the chromosome 4, co-segregated with one of the resistance genes. RM6506 was 11.0cM in the upper of the gene, while RM273 and RM252 were 6.0cMand 10.4cM below the gene, respectively (Figures 1 and 2). It has been reported that there were two BPH resistance genes, bph12(t) and Bph15(t) in the chromosome4. The bph12(t) and Bph15(t), both derived from O. Officinalis (Wall. ex Watt) with CC genome, have been located at around 56.2cM and 18.3cM of the Cornell rice map (McCouch et al., 2002), respectively, and the latter is a dominant gene (Yang et al., 2002; 2004). In this study, the BPH resistance gene is located at around88cM of the Cornell map (Figure 2), far from the locations of bph12(t) and Bph15(t). Therefore, this gene is most possible a new gene and tentatively designated as bph18(t).
|
|
|
|
The other resistance gene in 2183 was mapped in the chromosome 12, near the end of long arm, with genetic distance 16.7cM to the SSR marker RM17 (Figure 3). But the exact location of this locus has not decided because other markers for it have not been found. Further study for location of this gene is going on. Up till now, 4 BPH resistance genes, Bph1 (Hirabayashi and Ogawa, 1995), bph2 (Murata et al., 1998a; Jeon et al., 1999), Bph9 (Murata et al., 2001) and Bph10(t) (Murata,1997) have been reported in the chromosome 12.The Bph1 was a dominant gene closely linked withbph2 and located at a region between 64.1~64.7cM ofthe Cornell map. The exact location of Bph9 is not known as it was mapped by RAPD markers (Murata et al., 2001), but it is a dominant gene. The Bph10(t) is also a dominant gene located at the near upper of Bph1.In this study, the new gene is located in the end region of the chromosome below 91cM of the Cornell map and with at least 28cM distance to the other resistance gene in the same chromosome. Therefore, it is possibly a new gene and tentatively designated as bph19(t).
|
|
This research results, together with our former study in which another two new loci conferring BPH resistance were found (Yang et al., 2005), indicated that there were rich resistant sources to the BPH present in Guangxi common wild rice and these new genes would strongly help to the diverse utilization of resistance genes against the BPH.
5 Resistant Germplasm Enhancement
A lot of resistant germplasms were bred so as to create a large number of breeding materials with genetic diversity. A total of 293 crosses, backcrosses and multi-crosses have been made, from which 143 genetic stable lines with BPH resistance were obtained. The BPH resistance in the new breeding lines was compared with that of the parents. The mean resistance score to the biotypes 2 and Bangladesh in the parent 94-42-5-1were 2.86 and 2.63 respectively, and in the progeny lines were 2.2~3.0 and 1.3~3.0 respectively (Table 5).This indicated that the BPH resistance could be maintained after the gene transfer. As both the species possess the same AA genome, the transfer of the resistance genes does not meet difficulty.
|
|
6 Yield Performance of the BPH Resistant Breeding Lines
A few superior BPH resistant breeding lines and hybrids were evaluated for their yield potential. These lines showed reasonable higher yield as compared with the local hybrid. The highest yield of T5S/99-BPH71 reached 8 370kg/hm2, and a quality line 99Q736 could yield 6 045kg/hm2 (Table 6). This indicated the utilization of the BPH resistance derived from the wild rice O. rufipogon have made good progress in rice breeding.
|
|
Acknowledgements
This research project is supported by China National Natural Science Foundation (30360053), Science and Technology Department of Guangxi (Project No.Gui ke Neng 05112001-1A1) and Guangxi Key Laboratory of Subtropical Bioresource Conservation and Utilization. We also heartfully thank Professors Li Qing, Luo Shanyu, Wu Miaoxin and Li Daoyuan for their great technical contribution to the work.
References
Hirabayashi H., and Ogawa T., 1995, RFLP mapping of Bph- 1 (brown planthopper resistance gene) in rice, Breed Sci., 45: 369-371
Hirabayashi H., Angeles E.R., Kaji R., Ogawa T., Brar D.S., and Khush G.S., 1998, Identification of brown planthopper resistance gene derived from O. officinalis using molecular markers in rice, Breed Sci., 48(Suppl.1): 82
Jeon Y.H., Ahn S.N., Choi H.C., Hahn T.R. and Moon H.P., 1999, Identification of a RAPD marker linked to a brown New bred lines planthopper resistance gene in rice, Euphytica, 107: 23-28 Kabir M.A., and Khush G.S., 1988, Genetic analysis of resistance to brown planthopper in rice (Oryza sativa L.), Plant Breed., 100(1): 54-58
Kawaguchi M., Murata K., Ishii T., Takumi S., Mori N., and Nakamura C., 2001, Assignment of a brown planthopper (Nilaparvata lugens St!l) resistance gene bph4 to the rice chromosome 6, Breed. Sci., 51: 13-18 doi:10.1270/jsbbs.51.13
Khush G.S., Rezaul Karim A.N.M., and Angeles E.R., 1985, Genetics of resistance of rice cultivar ARC 10550 to Bangladesh brown planthopper biotype, J. Genet., 64(2-3): 121-125 doi:10.1007/BF02931140
Kinoshita T., 1995, Report of the committee on gene symbolization, nomenclature and linkage groups, Rice Genet. Newsl., 12: 9-153
Li Q., Luo S.Y., Shi A.X., Wei S.M. and Huang F.K., 1997, The biotypes of brown planthopper [Nilaparvata lugens (St"l)] with a view to its control, Act. Entom. Sin., 40 (suppl.): 139-146
Liu G.Q., Yan H.H., Fu Q., Qian Q., Zhang Z.T., Zhai W.X., and Zhu L.H., 2001, Mapping of a new gene for brown planthopper resistance in cultivated rice introgressed from Oryza eichingeri, Kexue Tongbao (Chinese Science Bulletin), 46(17): 1459-1462
Manisegaran S., Gopalan M., and Mohamed Hanifa A., 1993, Differential reaction of selected rice cultivars to brown planthopper, Nilaparvata lugens St#l, Indian J. Plant Protection, 21: 31-33
McCouch S.R., Teytelman L., Xu Y.B., Lobos K.B., Clare K., Walton M., Fu. B.Y., Maghirang R., Li Z.K., Xing Y.Z., Zhang Q.F., Kono I., Yano M., Fjellstrom R., DeClerck G., Schneider D., Cartinhour S., Ware D., and Stein L., 2002, Development and mapping of 2 240 new SSR markers for rice (Oryza sativa L.), DNA Res., 9(6): 199-207 doi:10.1093/dnares/9.6.199 PMid:12597276
Medina E.B., Bernal C.C., and CohenM.B., 1996, Role of host plant resistance in successful control of brown planthopper in Central Luzon, Philippines, InternationalRiceRes.Notes, 21: 53
Murata K., Fujiwara M., Kaneda C., Takumi S., Mori N., and Naka-mura C., 1998a, RFLP mapping of a brown planthopper (Nilaparvata lugens St$l) resistance gene bph2 of indica rice introgressed into a japonica breeding ine‘Norin-PL4’, Genes Genet. Syst., 73: 359-364 doi:10.1266/ggs.73.359
Murata K., Fujiwara M., Murai H., Mori N., Takumi S., and Naka-mura C., 1998b, Detailed mapping of brown planthopper resistance gene BPH9, by RAPD analysis, Breed. Res., 48(Suppl.1): 83
Murata K., Fujiwara M., Murai H., Takumi S., Mori N., and Nakamura C., 2001, Mapping of a brown planthopper (Nilaparvata lugens St%l) resistance gene Bph9 on the long arm of rice chromosome 12, Cereal Res. Commun., 29: 245-250
Murata K., Nakamura C., Fujiwara M., Mori N., and Kaneda C., 1997, Tagging and mapping of brown planthopper resistance genes in rice, In: Su J.C.(ed.), Proceed. 5th Intern. Symposium on Rice Molecular Biology, Yi-Hsien Pub., Taiwan, China, pp.217-231
Nomoto H., Ikeda R., and Kaneda C., 1989, New genes for resistance to brown planthopper, Nilaparvata lugens St&l, in rice, Jpn. J. Breed., 39: 23-28
Pathak P.K., and Lal M.N., 1976, Studies on varietal resistance to the brown planthopper, Nilaparvata lugens (St’l) and its biotypes, International Rice Res. Newsltt., 1: 8
Renganayaki K., Fritz A.K., Sadasivam S., Pammi S., Harrington S.E., McCouch S.R., Kumar S.M., and Reddy A.S., 2002, Mapping and progress toward map-based cloning of brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated rice, O. sativa, Crop Sci., 42(6): 2112-2117 doi:10.2135/cropsci2002.2112
Sidhu G.S., and Khush G.S., 1978, Genetic analysis of brown planthopper resistance in twenty varieties of rice, Oryza sativa L., Theor. Appl. Genet., 53: 199-203 doi:10.1007/BF00277368
Wu C.J., Jiang G.H., Li X., Liu T., Xu C.G., and He Y.Q., 2005, Dynamic detection and analysis of QTL for resistance to the brown planthopper, using a doubled-haploid rice population, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 3 (4): 456-462
Yang H., You A., Yang Z., Zhang F., He R., Zhu L., and He G., 2004, High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.), Theor. Appl. Genet., 110(1): 182-191 doi:10.1007/s00122-004-1844-0 PMid:15549231
Yang H.Y., Ren X., Weng Q.M., Zhu L.L. and He G.C., 2002, Molecular mapping and genetic analysis of a rice brown planthopper (Nilaparvata lugens St(l) resis tance gene, Yichuan (Hereditas), 136: 39-43 doi:10.1034/j.1601-5223.2002.1360106.x PMid:12184487
Yang L., Li R.B., and Li Y.R., 2005, Preliminary mapping the genes conferring resistant to brown planthopper (Nilaparvata lugens) in rice, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 3(6): 807-809
. PDF(1261KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Rongbai Li
. Lishu Li
. Sumei Wei
. Yanping Wei
. Yingzhi Chen
. Delang Bai
. Lang Yang
. Fengkuan Huang
. Weili Lu
. Xiangjun Zhang
. Xiaoyong Li
. Xinqing Yang
. Yuanwen Wei
Related articles
. Oryza rufipogon
. Oryza sativa
. Brown planthopper ( Nilaparvata lugens Stal)
. Resistant varieties
. Gene mapping
. bph18(t)
. bph19(t)
Tools
. Email to a friend
. Post a comment
.png)
.png)